In Foldit, there are two ways to mutate segments manually, in the game interface.
One version automatically picks the best amino acid for all segments (or selected segments). The second version mutates one segment (or selected segments) to a specific amino acid picked by the user.
These tools work slightly differently in the original and selection interfaces.
Original Interface[]

Mutate tool Actions menu (original interface)
In the original Foldit interface, the automatic version of mutate is available from Actions menu. This tool automatically mutates all mutable segments in the protein. The keyboard shortcut lowercase "m" can also be used to select this tool. The tool is available in all modes (pull, structure, note, and design).
Design mode (original interface)
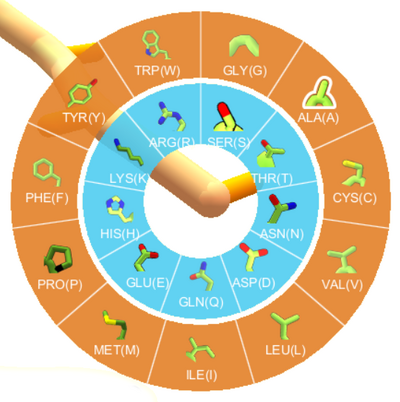
Mutate wheel (design mode, original interface)
In design mode, left-clicking on a segment brings up a wheel showing the amino acids. The hydrophobic amino acids are shown with orange backgrounds on the outer edge of the wheel. The hydrophilic amino acids are shown with blue backgrounds on the inner hub of the wheel. The desired amino acid can be selected by clicking on it, or typing its one-letter code (lowercase), as shown on the wheel. It's only possible to mutate one segment at a time. There is no keyboard shortcut for displaying the mutate options.
Not all possible amino acids are valid in all puzzles. For example, many recent design puzzles don't allow any cysteine segments. Puzzles may also disallow certain amino acids in certain positions. In general, some amino acids may be "greyed out" in the wheel, and clicking them has no effect. If you type the keyboard shortcut for one of the dimmed selections, the message "Selection contains invalid mutations" is displayed in the main window. (The same message is used in the selection interface.)
Selection Interface[]
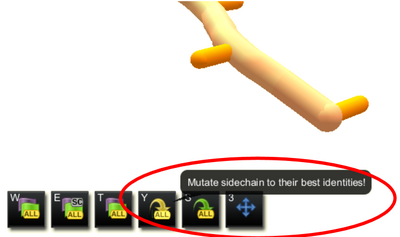
Mutate all option with no segments selected (selection interface)
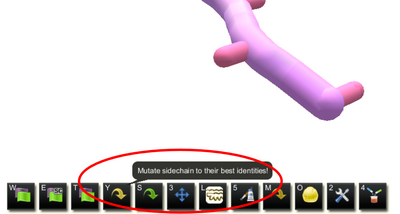
Mutate tool with segments selected (selection interface)
In the selection interface, there are two versions of automatic mutate. If no segments are selected, the mutate icon shows the word "all". Clicking the icon mutates all mutable segments in the protein. If one or more segments are selected, the word "all" disappears, and the tool mutates only the selected segments. The keyboard shortcut lower ase "y" can also be used to run both versions of the automatic mutate tool.
Mutate specific (selection interface)

Mutate specific menu (selection interface)
The selection interface can also mutate to a selected amino acid. If one or more segments are selected, another mutate tool appears in the horizontal tool bar. Clicking this icon or using the lowercase "m" keyboard shortcut displays an amino acid menu, similar to the wheel in the original interface, but arranged in rows. Clicking one of the amino acids or typing its one-letter abbreviation (lowercase) mutates all selected segments to the specified amino acid.
As with the original interface, not all amino acids are valid in all puzzles at all positions. Some amino acids may be "greyed out" for some selections. If you type the keyboard shortcut for one of the dimmed selections, the message "Selection contains invalid mutations" is displayed in the main window.
Historical version[]
This section contains information from a previous version of this page.
Screenshot - Mutate Sidechains
Mutate sidechains (hotkey "m" on the keyboard) automatically seeks out better proportioned sidechains for the backbone all at once. Locked backbone segments will not mutate. Available for design puzzles only.
Pestering voids won't get out of your way? Score goes up as you pack in that space! Shake things up a bit by trying Mutate Sidechains.
How does it work?[]
When I ask Foldit to mutate my protein, how does Foldit determine the structure? My guess is that if I select 4 residues, and mutate them to any amino acid type, then Foldit will not compute the energy of each of the 20*20*20*20 = 20^4 structures, and pick the best. I think there is some special search strategy so that it takes less time to find the best scoring structure.
The attribution of the following paragraph is unclear. It seems to be quoting someone in-the-know at Foldit central.
In answer to your question, we use a different search strategy than complete enumeration of the side chain identities. You are correct that it is very computationally demanding to find the best mutations using complete enumeration. To circumvent this cost, we use something called a 'Monte Carlo' search strategy to find the best combination of mutations (see wikipedia "Monte Carlo method" for an overview). The disadvantage is that we are never sure that the mutations chosen are the absolute 'best'.
The original paper describing the Monte Carlo search strategy for the Foldit system is described in: Brian Kuhlman and David Baker, PNAS (2000) vol.97 page 10383.