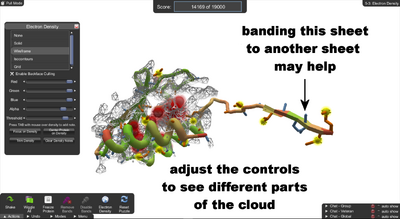
Electron Density. The Electron Density panel is displayed. Changing the settings on this panel changes the appearance of the electron density cloud.
In an Electron Density puzzle, the neat "guide" seen in Quest to the Native puzzles is replaced by a fuzzy "cloud" of electron density data gathered from x-raying the protein.
Madde's "Electron Density" video
Loci's "Electron Density" video
Just as in the QTTN puzzle, the goal is to fit the protein into the cloud.
(Here "density" means the probability of an electron being present, which is all the x-rays can tell you. The denser the probability cloud, the higher the chances are that there's a big atom nearby.)
On the Actions menu in the lower left, there's an Electron Density button, which opens the Electron Density control panel. (You can also use shift+E to toggle the panel.)

Banding the stray sheet. The protein has been rotated and the electron density view set to "none". Also, display of clashes, exposeds, and voids have been toggled off (shortcuts shift+C, shift+X, and shift+V).
The Electron Density panel controls the appearance of the cloud. (You can also hide the cloud by selecting "None", sometimes the cloud is too much information.)
The "Center Protein on Density" button on the ED panel is similar to "Align Guide" in the QTTN puzzle. It moves the protein into the density, but it doesn't change the shape of the protein, and it doesn't try to figure out the best alignment.
The color sliders on the ED panel let you change the color of the cloud, and the Alpha slider changes the brightness of the cloud.
The Threshold slider controls how much of the density is shown. If the Threshold slider is all the way to the left, the most density is shown. This includes areas where there's only a low probability of an electron. As the slider moves to the right, the cloud gets smaller, showing only higher probabilities of an electron being present.

Electron Density, solved. Big rings, like the phenylalanine on the right, are often the easiest to find in the cloud.
In this puzzle, the protein has a sheet that's out of line. To get started, shake the protein, then draw bands to pull the wayward sheet toward the others. You may want to add bands or use freezes to hold the other sheets in place. Then wiggle to pull the sheet into alignment.
Just fixing the stray sheet may be enough to solve this puzzle. But try experimenting with the sliders and views to see how the cloud changes. Clicking "Center Protein on Density" a time or two may also help.
There are several keyboard shortcuts that may help. Try hiding exposeds using shift+E, and hiding voids using shift+V.
You may want to show all sidechains using shift+A, since the shapes of sidechains can be seen in the cloud. Hide all sidechains with shift+D or go back to showing stubs of sidechains with shift+T.
If you hover over a part of the protein and hit shift+Q, the view changes to allow you to see that part better. A lowercase "q" restores the view of the whole protein.
PDB entry 1HDN is the same protein seen in the electron density puzzle.
Technical stuff: electron density puzzles are one of the toughest challenges in Foldit. They're very close to what real scientists do. Sometimes automatic methods can figure out the correct shape of a protein from the x-ray results. Sometimes, the automated methods fail, and the scientists have to figure it out. See Electron Density Puzzles for more.
Unlike many of the other intro puzzles, this puzzle is a complete protein. It's been studied and "solved" several times. The electron density cloud seen in this puzzle is probably taken from one of these studies.
Information about solved proteins is usually deposited in the Protein Data Bank or PDB at rcsb.org. Proteins in the PDB have a four-character ID. PDB entry 1HDN is a good example of the protein in this electron density puzzle.
The same sequence of amino acids (or primary structure) can also be found in PDB entries 3EZE, 3EZB, 3EZA, 2XDF, 2LRL, 2LRK, 2JEL, 1VRC, 1POH, 1PFH, 1J6T, and 1GGR. In most of these other entries, the protein from this puzzle is shown combined with other, separate pieces. The pieces are called chains, are identified as A, B, C, and so on. PDB 1HDN has the puzzle protein as chain A, the only chain. In PDB 2JEL, the protein from this puzzle doesn't show up until chain P!
The protein is a component of phosphotransferase, an enzyme that bacteria use to process sugars.

